Cellular Biochemistry of Multienzyme Metabolic Complexes in Living Cells: Metabolons
 The broad objective of my laboratory is to contribute to our understanding of how sequential metabolic enzymes are organized in living cells and how such metabolic organizations are spatially and/or temporarily regulated in response to cellular signals relevant to human diseases; including but not limited to cancer, diabetes and obesity.
The broad objective of my laboratory is to contribute to our understanding of how sequential metabolic enzymes are organized in living cells and how such metabolic organizations are spatially and/or temporarily regulated in response to cellular signals relevant to human diseases; including but not limited to cancer, diabetes and obesity.
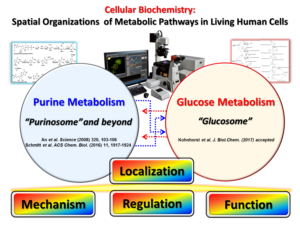
Since the concept of the metabolic complex was conceived in 1970s, the discovery of the “purinosome”, a reversible metabolic macromolecular assembly in human cells [An et al. Science (2008) 320, 103], stands as the first example of transient metabolic assemblies found in living cells. Recently, we have identified that a scaffold enzyme of the purinosome can be spatially organized into its own self-assembly (i.e. FGAMS self-assembly), by which de novo purine biosynthesis is downregulated in HeLa cells [Schmitt et al. ACS Chem. Biol. (2016) 11, 1917]. In addition, we have demonstrated that the core complex of the purinosome, which is formed by 3 enzymes catalyzing the first half of de novo purine biosynthesis, can be independently regulated by signaling pathways [Schmitt et al. PLOS One (2018)13, e0195989]. In parallel, we have also identified a functionally relevant, multienzyme metabolic complex for glucose metabolism, the “glucosome”, in living human cells [Kohnhorst et al. J.Biol. Chem. (2017) 292, 9191]. All of these discoveries thus set the stage for an extraordinary opportunity to explore the spatial and temporal advantages to the cell in assembling and disassembling this cluster of metabolic enzymes. Indeed, my laboratory at UMBC is highly motivated to explore the spatial assemblies of de novo purine biosynthesis and glucose metabolism as the heart of human disease mechanisms.
Collectively, the research projects in my laboratory have potentials to invoke a paradigm shift in our thinking about the operation of cellular biosynthetic pathways, and outcomes will be beneficial for human health.
Funding from NIH-NIGMS (R01), NIH-NCI (R03), AACR and UMBC.
